方法分享--空间转录组(visium等)提升分辨率
原创方法分享--空间转录组(visium等)提升分辨率
原创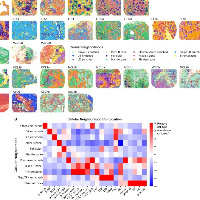
追风少年i
修改于 2026-01-08 16:08:21
修改于 2026-01-08 16:08:21
作者,Evil Genius
2026赛季这就算开打了,谁可以在单细胞空间领域拔得头筹?

这里面超过一半的公司我都没听过,主要也是做产品的公司,还是那样,产品具有不可替代性,想做单细胞空间,必须用仪器设备试剂获得数据,这样就可以不断地迭代,生物信息分析比较低端,还是不如做产品好。
2026的终极目标,就是活下去,我妈找人算命,说我走背字要4-6年才能转运,就算最快的4年,也要到2027年好转了,以前我从来不信命,现在的我怀疑这个世界都是虚拟的,挺过2026,我相信会时来运转。
2026开始了,那么25年之前搜集的文献也就算完成了,新的文献将会汇总到2026年,每年的文献我都有好好整理。
其中2023年文献
2024年文献
2025年文献

2025的培训,主要也是针对visium、HD、Xenium、CosMx、stereo-seq 的,空间方向,更能体现产品的价值,visium/HD现在大约3万/样本,Xenium、CosMx就更贵了,大panel都上了10万,不得不说,数据分析的价值比起产品真的太低了。
关于2025的培训,大家自行参考, 基本上认真学一遍就成,而且不像产品一样具有迭代性,学会了也就完了,产品是做一个付一次费用,技能是学一遍也就不需要再用了。
还是要认真做产品才好。
生信主要有两个方向,算法方向和应用方向,像我这样天天分析空间项目的就是应用方向。
今天我们来分享方法,低精度空转的分辨率提升,参考文献如下,看作者介绍,应该中美合作的文章。
知识积累
主流平台(如10x Visium)的空间分辨率仍较有限——每个测序“点”(spot)通常包含多个细胞,且组织中存在大量未被覆盖的区域。虽然Stereo-seq、HD等新技术提升了分辨率,但其数据更加稀疏和嘈杂,常需将多个spot合并为“meta-spot”以提高信噪比,却牺牲了空间精度。
ST实验通常配有高分辨率组织学图像(如H&E染色或免疫荧光图像),可提供更精细的组织形态与细胞排布信息。
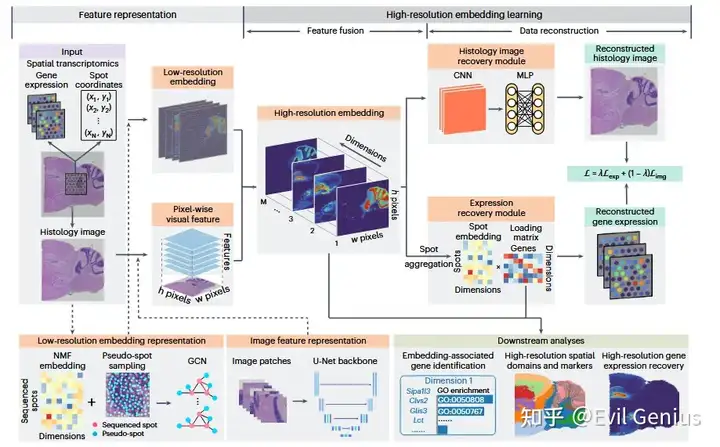
结果1、SpaHDmap 融合基因表达数据与组织学图像,实现高分辨率的降维
SpaHDmap 框架包含两个主要组成部分:特征表示(feature representation)。
在特征表示模块中,SpaHDmap 分别从空间转录组(ST)数据中学习一个低分辨率嵌入,并从组织学图像中提取逐像素的视觉特征。
低分辨率嵌入首先通过对基因表达数据应用非负矩阵分解(NMF),为已观测到的测序点(spots)生成初始嵌入;随后,通过一个图卷积网络(GCN)对该嵌入进行去噪并扩展至整个组织区域。该GCN的节点不仅包括实际观测到的spots,还包括用于提升空间覆盖度的“伪点”(pseudo-spots),并融合了组织学图像信息。
对于图像特征表示,SpaHDmap 训练一个多通道 U-Net 网络以重建图像块(image patches),然后将其主干网络(backbone)迁移用于提取逐像素的视觉特征。
随后,在高分辨率嵌入学习模块中,SpaHDmap 采用一个多模态融合的编码器–解码器架构,通过同步重建ST基因表达和组织学图像来学习高分辨率嵌入:
编码器(即特征融合模块)将上述低分辨率嵌入与逐像素图像特征融合,并映射到一个高分辨率嵌入空间;
解码器(即数据重建模块)则基于所学嵌入,分别重建基因表达和组织学图像。
与NMF类似,嵌入向量以及表达重建模块中的可学习加载矩阵(loading matrix)的所有元素均被约束为非负值,从而确保得到一种可解释的、基于“部分组合”的高分辨率数据表示。
整体重建损失是基因表达重建损失与组织学图像重建损失的加权和。
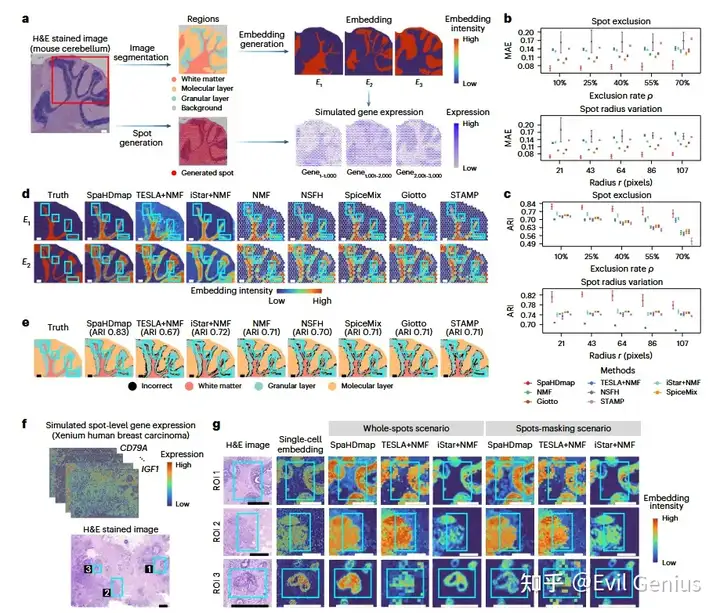
小鼠脑的分析效果
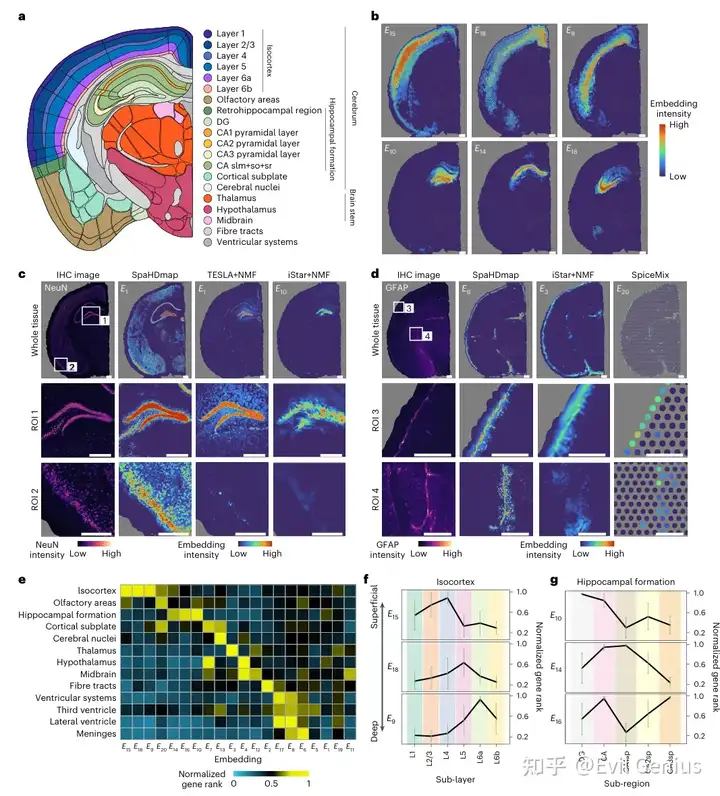
SpaHDmap 在小鼠脑数据集中可解释的高分辨率嵌入
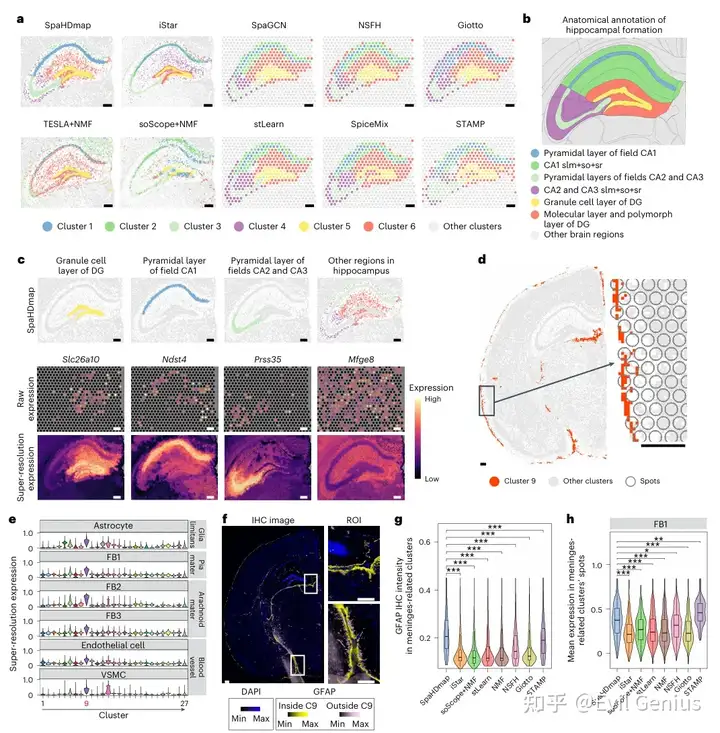
SpaHDmap 在小鼠脑数据中识别出精细的空间结构
SpaHDmap 在多个髓母细胞瘤样本中成功恢复了疾病状态特异性的高分辨率嵌入
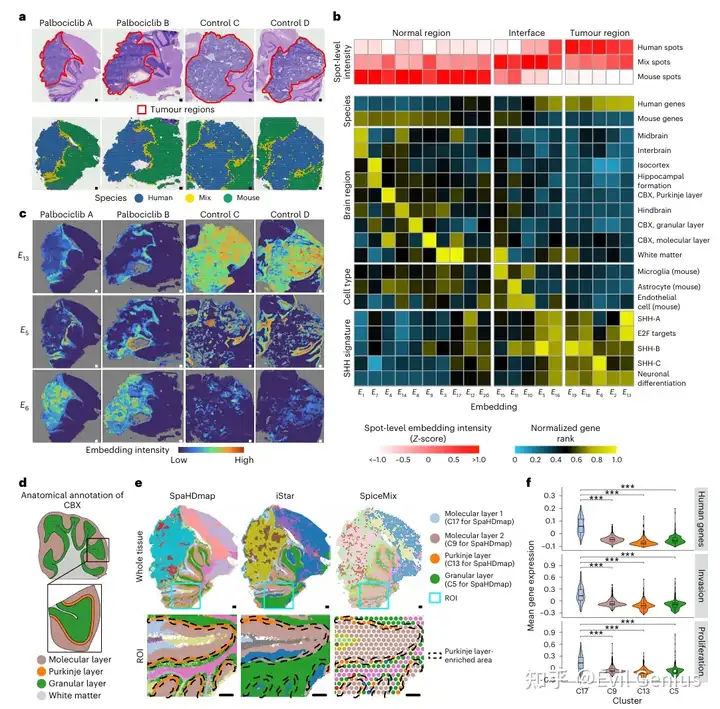
当然,文章还展示了很多其他的示例
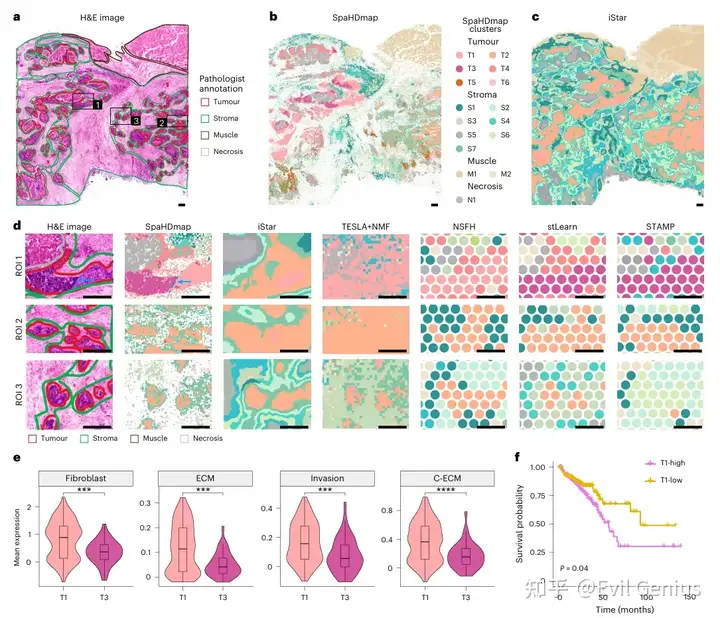
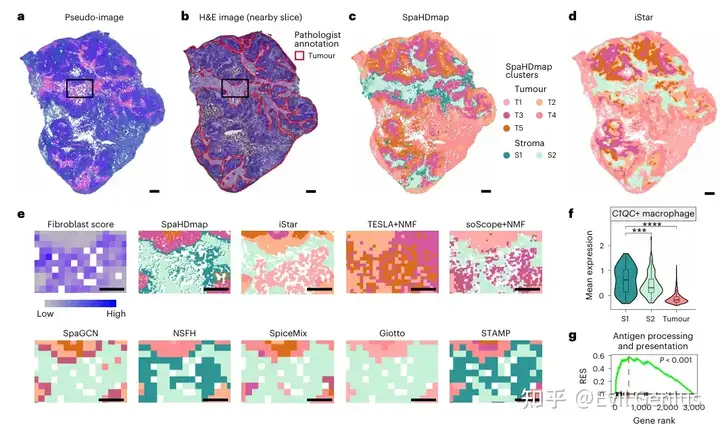
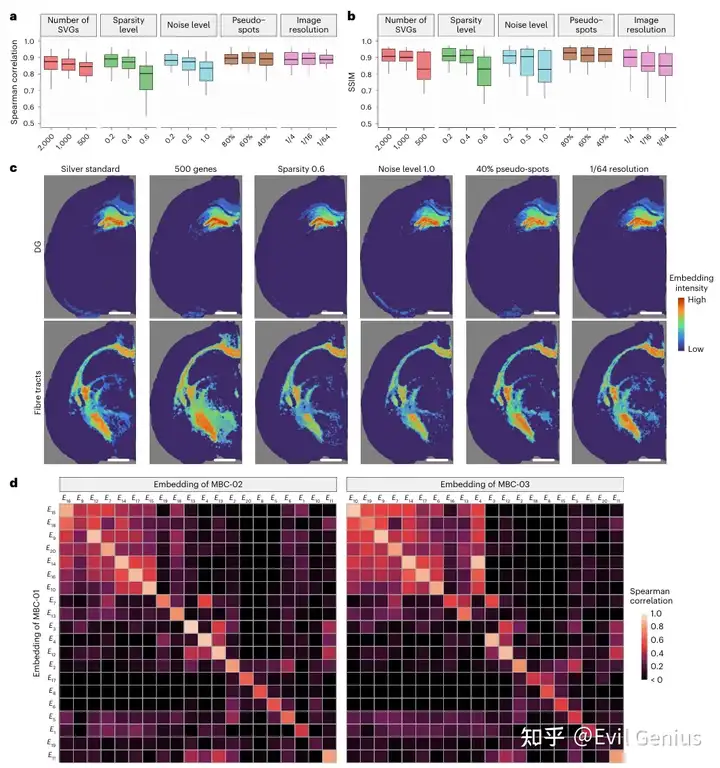
最后,我们来看看代码
import torch
import numpy as np
import scanpy as sc
import SpaHDmap as hdmap
rank = 20
seed = 123
verbose = True
np.random.seed(seed)
torch.manual_seed(seed)
root_path = '../experiments/'
project = 'MPBS01'
results_path = f'{root_path}/{project}/Results_Rank{rank}/'
section_id = 'V1_Mouse_Brain_Sagittal_Posterior'
# Download the data from the 10X website (set include_hires_tiff=True to download the hires image)
adata = sc.datasets.visium_sge(section_id, include_hires_tiff=True)
image_path = adata.uns["spatial"][section_id]["metadata"]["source_image_path"]
# or load the data from a local folder
# adata = sc.read_visium(f'data/{section_id}', count_file='filtered_feature_bc_matrix.h5')
# image_path = f'data/{section_id}/image.tif'
# Load the data from the 10X Visium folder
mouse_posterior = hdmap.prepare_stdata(adata=adata,
section_name='mouse_posterior',
image_path=image_path,
scale_rate=1)
hdmap.select_svgs(mouse_posterior, n_top_genes=3000, method='moran')
mouse_posterior.show()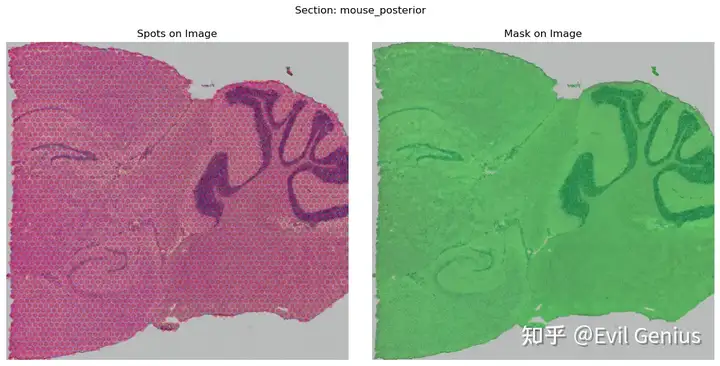
# Load the data from file paths
mouse_posterior = hdmap.prepare_stdata(section_name='mouse_posterior',
image_path='data/V1_Mouse_Brain_Sagittal_Posterior/image.tif',
spot_coord_path='data/V1_Mouse_Brain_Sagittal_Posterior/spatial/tissue_positions_list.csv',
spot_exp_path='data/V1_Mouse_Brain_Sagittal_Posterior/filtered_feature_bc_matrix.h5',
scale_rate=1,
radius=45) # Has to be provided if load from file paths
hdmap.select_svgs(mouse_posterior, n_top_genes=3000, method='moran')
###Run SpaHDmap
# Initialize the SpaHDmap runner
mapper = hdmap.Mapper(mouse_posterior, results_path=results_path, rank=rank, verbose=verbose)
# Run all steps in one function
mapper.run_SpaHDmap(save_score=False, save_model=True, load_model=True, visualize=True, format='png')
# If you want to lower the GPU memory usage, you can set `batch_size` to a smaller number
# mapper.args.batch_size = 16 (default 32)
# mapper.run_SpaHDmap(save_score=False, save_model=True, visualize=False)
# Run NMF on the gene expression data
mapper.get_NMF_score(save_score=False)
print(mouse_posterior.scores['NMF'].shape)
mapper.visualize(mouse_posterior, use_score='NMF', index=2)
# mapper.visualize('mouse_posterior', use_score='NMF', index=2) # visualize given the name
# mapper.visualize(use_score='NMF', index=2) # ignore the section name if only one section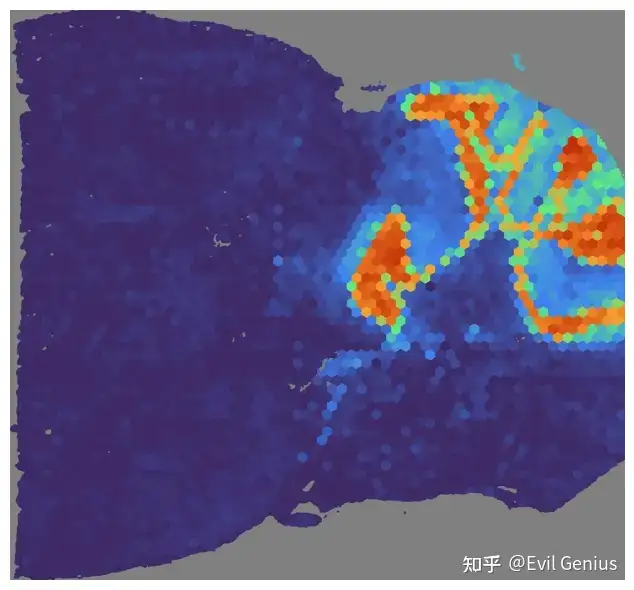
# Save all NMF scores into `results_path/section_name/NMF`
mapper.visualize(use_score='NMF', format='png') # default is 'png'
# Pre-train the SpaHDmap model via reconstructing the HE image
mapper.pretrain(save_model=True)
# Train the GCN model and get GCN score
mapper.get_GCN_score(save_score=False)
print(mouse_posterior.scores['GCN'].shape)
# Visualize the GCN score
mapper.visualize(mouse_posterior, use_score='GCN', index=2)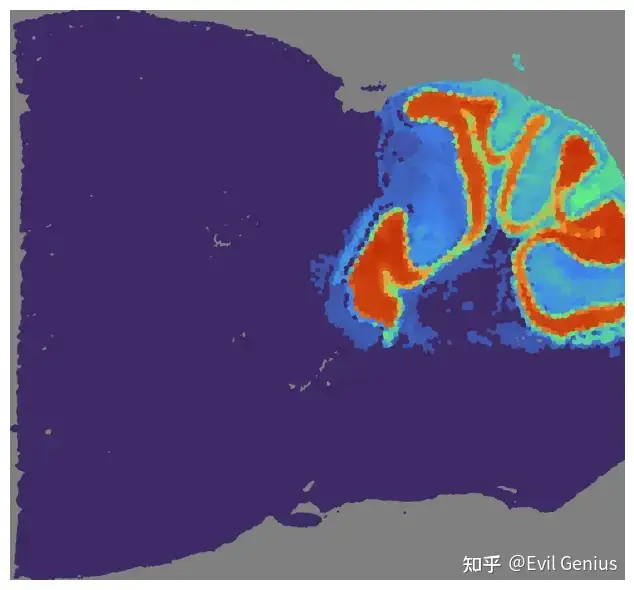
# Save all GCN scores into `results_path/section_name/GCN`
mapper.visualize(use_score='GCN', format='png')
# The refined metagene matrix based on the GCN score
print(mapper.metagene_GCN.shape)
# Get the VD score
mapper.get_VD_score(use_score='GCN')
# Train the SpaHDmap model
# If the model has been trained and saved, mapper will load the model directly
mapper.train(save_model=True, load_model=True)
# Get the SpaHDmap score
mapper.get_SpaHDmap_score(save_score=False)
print(mouse_posterior.scores['SpaHDmap'].shape)
# Visualize the SpaHDmap score
mapper.visualize(mouse_posterior, use_score='SpaHDmap', index=2)
mapper.visualize(use_score='SpaHDmap')
# Load the STData object
mouse_posterior = hdmap.prepare_stdata(st_path=(results_path + 'mouse_posterior.st'))
# Initialize the SpaHDmap runner with the loaded STData object
mapper = hdmap.Mapper(mouse_posterior, results_path=results_path, rank=rank, verbose=verbose)
# Cluster pixels based on the SpaHDmap score
mapper.cluster(use_score='SpaHDmap', resolution=0.8, show=True)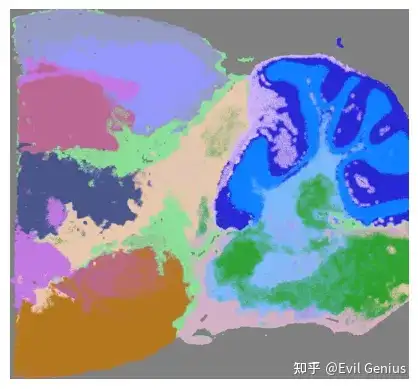
# Visualize the clustering results of SpaHDmap
mapper.visualize(target='cluster', use_score='SpaHDmap', format='pdf')
Recover high-resolution gene expression
mouse_posterior.genes[:5]
# Load the metagene matrix from the results path
mapper.load_metagene()
# Recover and visualize the high-resolution gene expression for a specific gene
mapper.recovery(gene=mouse_posterior.genes[:5], use_score='SpaHDmap')
mapper.visualize(gene='Pcp2')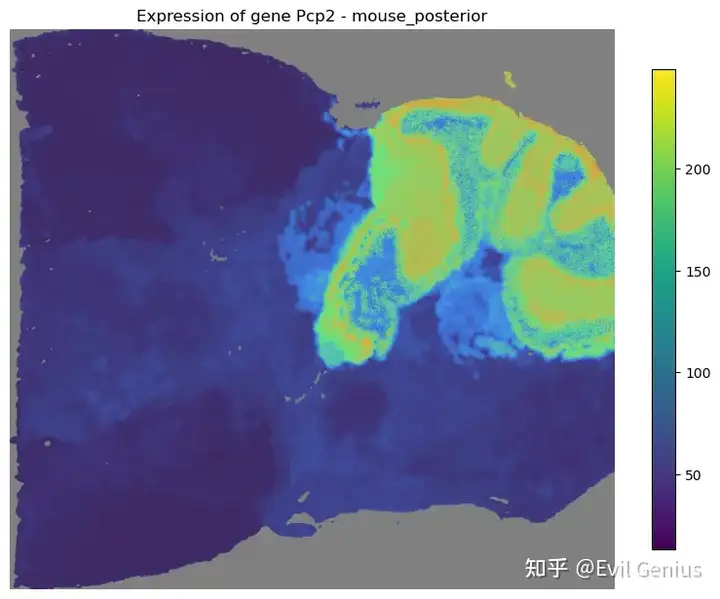
mapper.visualize(gene='Mbp')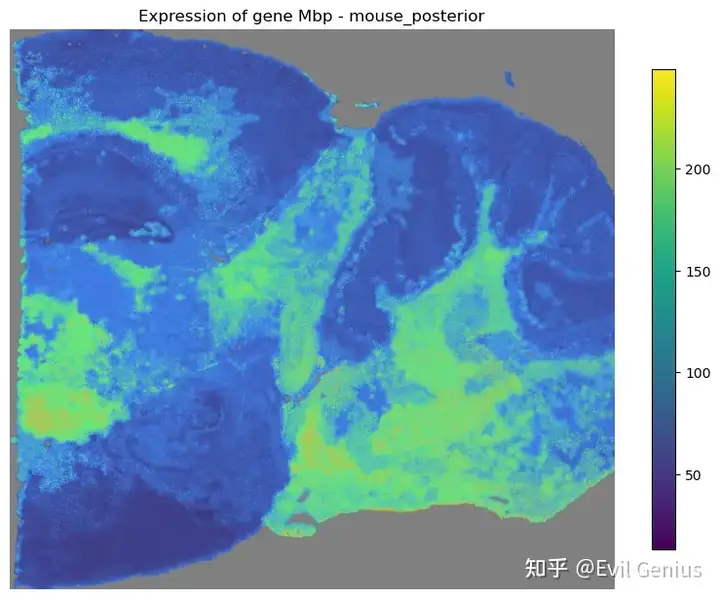
生活很好,有你更好。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
评论
登录后参与评论
推荐阅读
目录

