R tips:使用最近邻算法进行空间浸润带的计算
本文使用最近邻算法进行浸润带的计算。
空间组学中,有的时候需要对免疫浸润带进行特定距离的划分,形成一层一层的浸润区域。
以10X的官方xenium示例数据为例https://www.10xgenomics.com/datasets/ffpe-human-breast-with-pre-designed-panel-1-standard。
圈选ROI并计算浸润边界
下载的数据使用Xenium explorer打开,然后找到需要进行计算浸润带的位置,并根据方向将相应的全部选中。
如下图所示,假设中间的位置是需要进行浸润带计算的位置,而需要计算浸润带的方向是向下,则在Xenium explorer中选择套索工具仔细的圈画浸润边界,并将浸润带计算方向上的所有细胞选中。
然后在Xenium explorer中将图示的ROI区域的边界坐标下载下来。
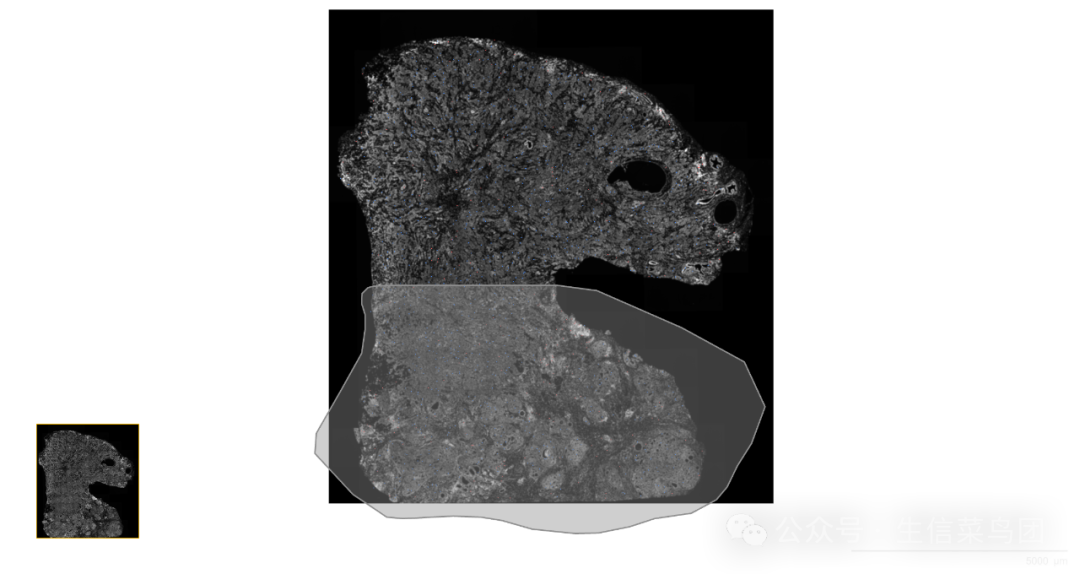
由于Xenium explorer无法圈画单条线,至少也是一个闭合区域,因此需要先计算好浸润边界的位置:
library(tidyverse)library(sp)# 读取ROI的边界坐标及细胞的质心坐标tumor_boundary <- read_csv("data/coordinates.csv", skip =2)cells_position <- arrow::read_parquet("Xenium_V1_FFPE_Human_Breast_IDC/cells.parquet")# 然后根据ROI边界坐标,将细胞分配到ROI中cell_idx <-sp::point.in.polygon(cells_position$x_centroid,# pointcells_position$y_centroid,# pointtumor_boundary$X,# polygontumor_boundary$Y-
)%>% -
as.logical # only 0 is FALSE, all others is TRUE # 于是tumor_area_1即是ROI中的细胞,图示中的所有细胞# tumor_area_2是剩余细胞,也就是图示中的上方未被框选中的细胞tumor_area_1 <- cells_position[cell_idx,]%>% mutate(x = x_centroid, y = y_centroid )tumor_area_2 <- cells_position[!cell_idx,]%>% mutate(x = x_centroid, y = y_centroid )
获得了浸润边界的两组细胞之后,就可以进行浸润边界的计算:
# 根据tumor_area_1和tumor_area_2找到绘制的tumor boundary# 寻找1和2中的最近邻点,以20um(大概一个细胞,根据效果调整)为限度,# 如果没有nn点则不是边界点。# 找到1和2中的边界点,取均值即是最终的边界点# dat: tumor_area_1# query: tumor_area_2nn_left_right <-RANN::nn2(tumor_area_1[2:3],tumor_area_2[2:3],k =10,searchtype ='radius',radius =20# 若边界未能找到,则调整radius的值-
) non_boundary_idx <- apply(nn_left_right$nn.idx ==0,1, all)if(sum(non_boundary_idx)== nrow(tumor_area_2)){-
print("radius is small, no boundary point.") }boundary_idx_1 <- nn_left_right$nn.idx[! non_boundary_idx,1]boundary_1 <- tumor_area_1[boundary_idx_1,]boundary_idx_2 <-! non_boundary_idxboundary_2 <- tumor_area_2[boundary_idx_2,]# comuted boudary coordsboundary_coords_df <-data.frame(- x = (boundary_1x_centroid + boundary_2x_centroid) / 2,
- y = (boundary_1y_centroid + boundary_2y_centroid) / 2
-
)
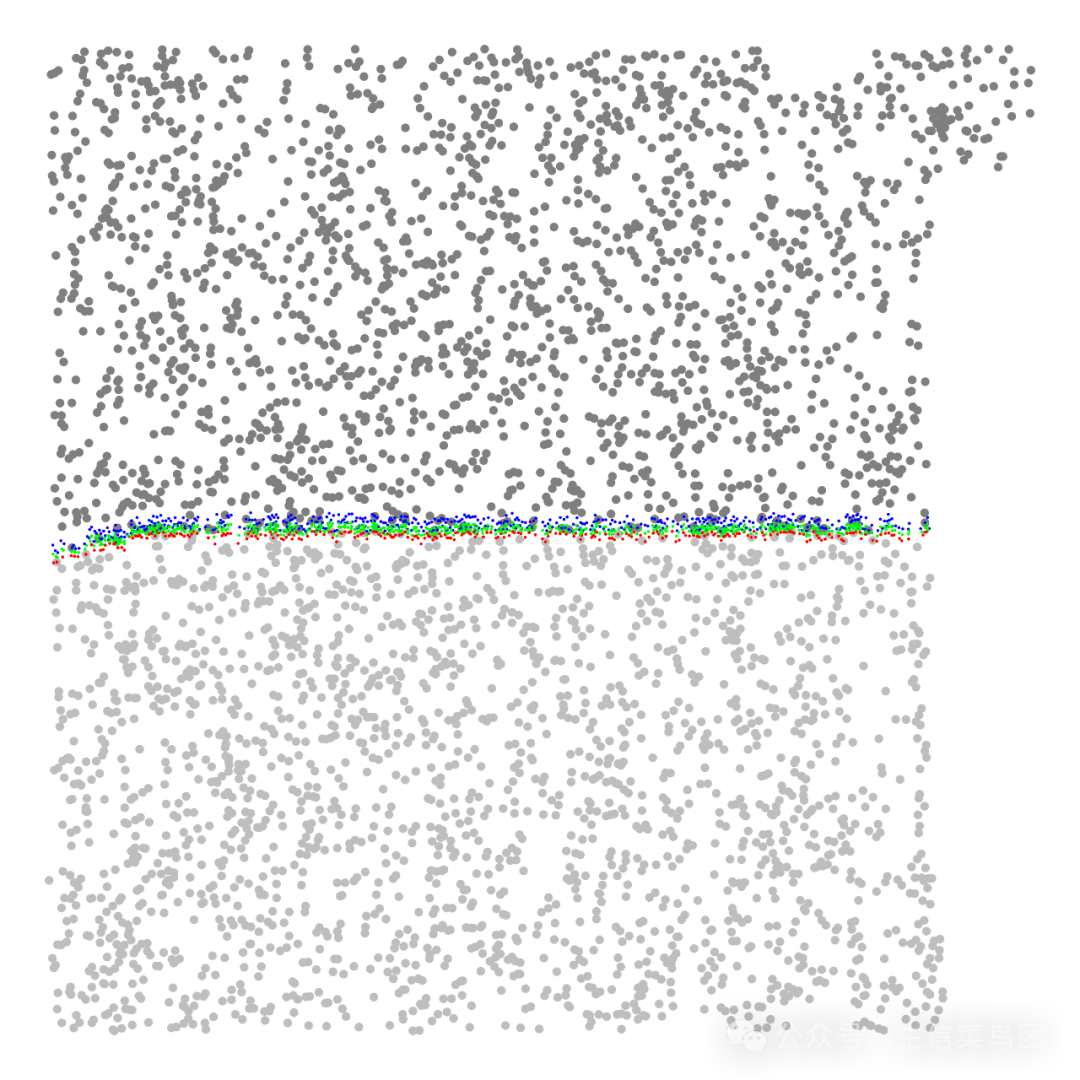
绘图展示,计算的圈画的浸润边界位置:
# 只展示浸润边界附近区域的细胞boundary_df_for_plot <-boundary_coords_df %>%-
.[chull(.),]%>% map(range)%>%map(~.x + c(-500,500))# 定义用于筛选细胞的函数filter_points <-function(dat, boundary){- x_idx <- datx_centroid < boundary
- y_idx <- daty_centroid < boundary
dat[x_idx & y_idx,]}# 绘图ggplot()+geom_point(-
# left area data = tumor_area_1 %>% dplyr::slice_sample(prop =0.1)%>% filter_points(boundary_df_for_plot),# 仅绘制10%的点,加快绘制速度aes(x = x_centroid, y =-y_centroid),color ="gray"-
)+ geom_point(-
# right area data = tumor_area_2 %>% dplyr::slice_sample(prop =0.1)%>% filter_points(boundary_df_for_plot),# 仅绘制10%的点,加快绘制速度aes(x = x_centroid, y =-y_centroid),color ="gray50"-
)+ geom_point(-
# comute left_boundary data = boundary_1,aes(x = x, y =-y),color ="red",size =0.1-
)+ geom_point(-
# compute right_boundary data = boundary_2,aes(x = x, y =-y),color ="blue",size =0.1-
)+ geom_point(-
# the final tumor boundary data = boundary_coords_df,aes(x = x, y =-y),color ="green",size =0.1-
)+ theme_void()
如下图所示,计算的浸润边界是绿色点,用于计算浸润边界的上下边界配对点是红蓝色点。
使用最近邻算法往下寻找浸润区域
假设需要以250um为单位,分别找到250um 500um及750um的浸润区域,则可如下操作:
先定义一个最近邻的工具函数:
# reduceFindNN find all (k) nn points in a certain radius,# k would be increased when k is not enough for a certain radiusreduceFindNN <-function(dat, k =10, query = NULL, searchtype ="radius", radius =100, mpp =1, max_k =2000){-
if(is.null(query)){ query <- dat-
} stopifnot(all(c('x','y')%in% colnames(dat)))stopifnot(all(c('x','y')%in% colnames(query)))dat <- dat[c('x','y')]query <- query[c('x','y')]nn_dat <- RANN::nn2(dat,query = query,k = k,radius = radius / mpp # radius, um -> pixel-
) - k_enough <- sum(apply(nn_dat
cat(str_glue("k is {k} and is enough for {k_enough * 100}% cells.\n\n"))-
while(k < max_k && k_enough <0.99){ -
if(k <700){ k <- k +100-
}elseif(k <2000){ k <- k +200-
}elseif(k <5000){ k <- k +500-
}else{ k <- k +1000-
} nn_dat <- RANN::nn2(dat,query = query,searchtype = searchtype,k = k,radius = radius / mpp # radius, um -> pixel-
) - k_enough <- sum(apply(nn_dat
cat(str_glue("k is {k} and is enough for {k_enough * 100}% cells.\n\n"))-
} -
return(nn_dat) }
浸润带的计算:
radius_ls <- c(250,500,750)%>% set_names(.,.)coords_tumor_infiltration_ls <-radius_ls %>%map(function(radius){dat_nn <-reduceFindNN(dat = tumor_area_1,# %>% rbind(tumor_boundary_coords) %>% distinct(),query = boundary_coords_df,# tumor_boundary_coords,radius = radius,mpp =1,k =100,max_k = nrow(tumor_area_1)# 2000-
) -
# all nn points is infiltration points points_idx <- dat_nn$nn.idx %>% c()%>%.[.!=0]%>% unique-
# infiltration area coords <- tumor_area_1[points_idx,]coords-
})
750um浸润带减去500um浸润带就是500-750um浸润带,以此类推:
# 当前浸润带减去上一层的浸润带coords_tumor_infiltration_ls2 <- list()for(i in rev(seq_along(coords_tumor_infiltration_ls))){name <- names(coords_tumor_infiltration_ls)[i]-
if(i ==1){ coords_tumor_infiltration_ls2[[name]]<- coords_tumor_infiltration_ls[[1]]-
return() -
} present_df <- coords_tumor_infiltration_ls[[i]]next_df <- coords_tumor_infiltration_ls[[i-1]]- coords_tumor_infiltration_ls2[[name]] <- present_df[!present_df
}# 获得浸润带 ----------coords_tumor_infiltration_df <-coords_tumor_infiltration_ls2 %>%enframe(name ="radius")%>%unnest(value)
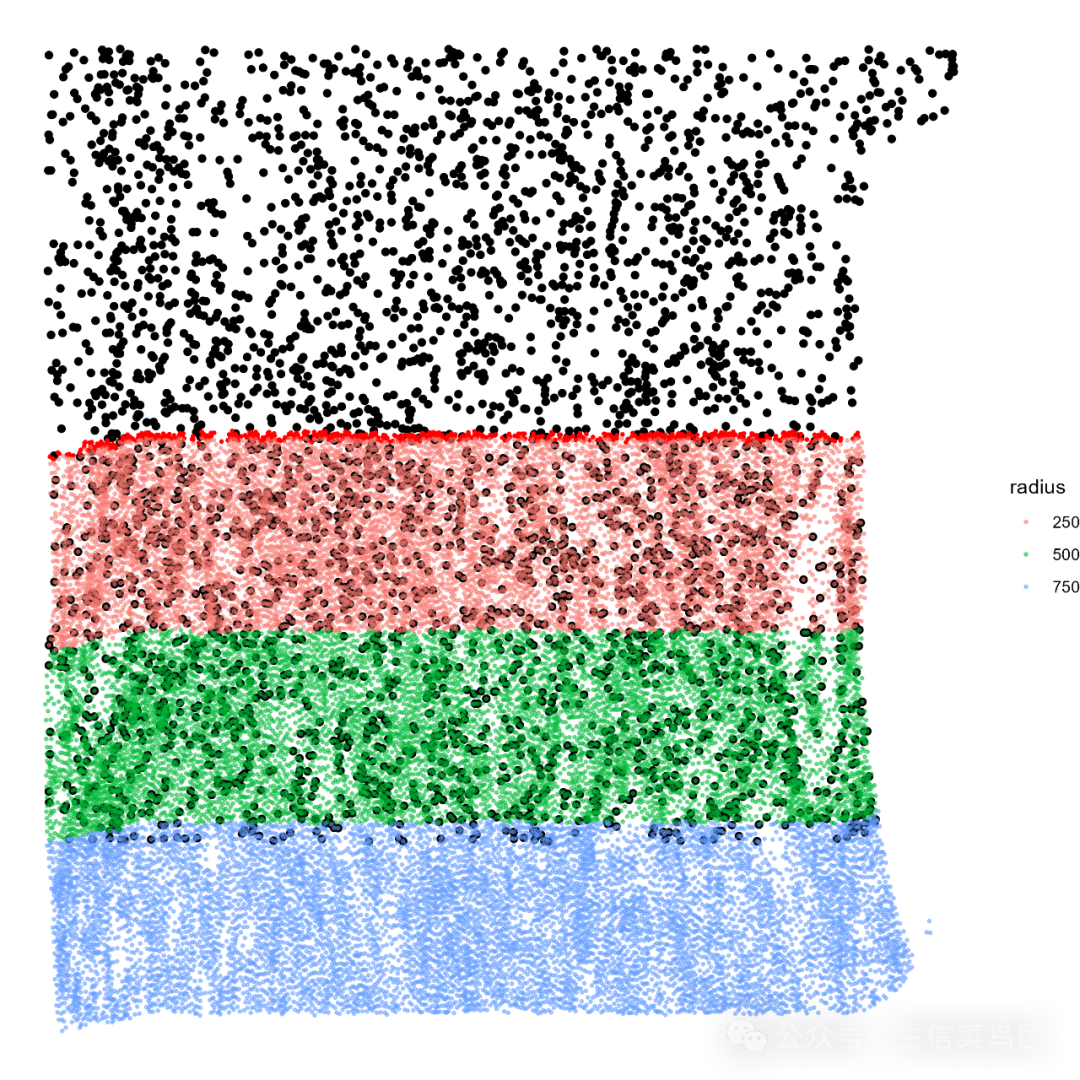
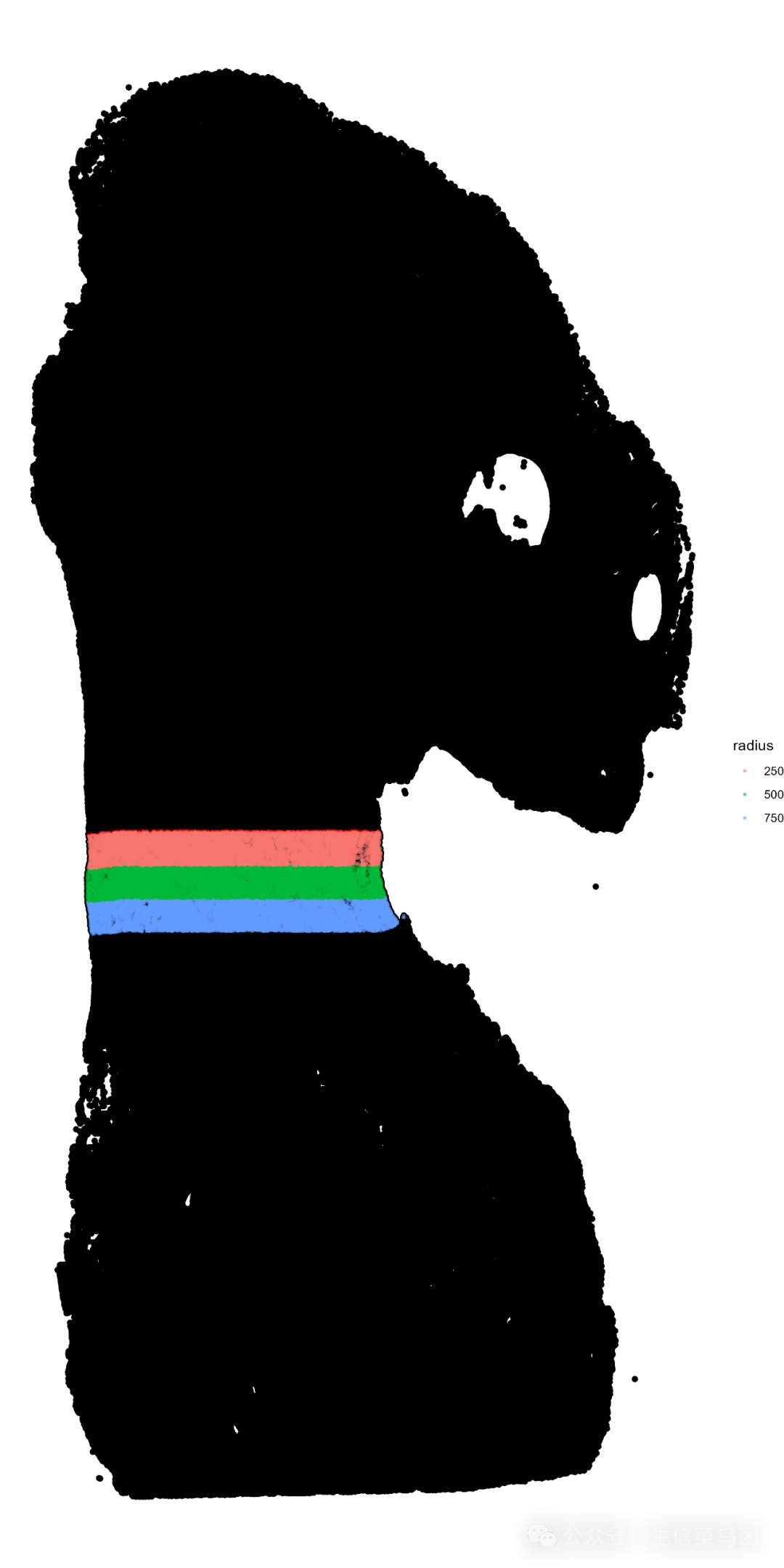
获得浸润区域如下:
p_infile_full <-cells_position %>%ggplot(aes(x = x_centroid , y =-y_centroid))+geom_point()+geom_point(data = boundary_coords_df,aes(x = x, y =-y),color ="red",size =0.5-
)+ geom_point(data = coords_tumor_infiltration_df,aes(x = x, y =-y, color = radius),-
# color = "blue", alpha =0.5,size =0.5-
)+ theme_void()p_infile_subset <-cells_position %>%dplyr::slice_sample(prop =0.1)%>%filter_points(boundary_df_for_plot)%>%ggplot(aes(x = x_centroid , y =-y_centroid))+geom_point()+geom_point(data = boundary_coords_df,aes(x = x, y =-y),color ="red",size =0.5-
)+ geom_point(data = coords_tumor_infiltration_df,aes(x = x, y =-y, color = radius),-
# color = "blue", alpha =0.5,size =0.5-
)+ theme_void()
如下图所示,浸润边界的浸润带展示:
全图展示的浸润带:
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-02-15,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

